Скрытая информация :: Авторизуйтесь для просмотра »
To observe relationships between features in our dataset, we could use a
scatter plot. A
scatter plot of the data puts one feature along the x-axis and another along the y-axis, and draws a dot for each data point. For scatter plotting how our data is distributed based on Sepal Length Width features, we could use the code below:
sns.FacetGrid(full_data,hue="Classification").map(plt.scatter,"sepal length (cm)",
"sepal width (cm)").add_legend()
plt.show()

To plot datasets with more than three features, we could use a pair plot, which looks at all possible pairs of features.
If you have a small number of features, such as the four we have here, this is quite reasonable.
You should keep in mind, however, that a pair plot does not show the interaction of all features at once, so some interesting aspects of the data may not be revealed when visualizing it this way.
We could use the
pairplot function in the
seaborn library as follows:
sns.pairplot(mydata)

Another way to do scatter plotting is to use scatter matrix existed in plotting module comes with pandas library. the following code creates a scatter matrix from the dataframe, and colors will be by Classes or labels:
from pandas.plotting import scatter_matrix
colors = list()
palette = {0: "red", 1: "green", 2: "blue"}
for c in np.nditer(iris.target): colors.append(palette[int(c)])
grr = scatter_matrix(mydata, alpha=0.3,figsize=(10, 10),
diagonal='hist', color=colors, marker='o', grid=True)

From the plots, we can see that the three classes seem to be relatively well separated using the sepal and petal measurements. This means that a machine learning model will likely be able to learn to separate them.
To show density of the length and width in the species, we could use
violin plot of all the input variables against output variable which is Species.
plt.figure(figsize=(12,10))
plt.subplot(2,2,1)
sns.violinplot(x="Classification",y="sepal length (cm)",data=full_data)
plt.subplot(2,2,2)
sns.violinplot(x="Classification",y="sepal width (cm)",data=full_data)
plt.subplot(2,2,3)
sns.violinplot(x="Classification",y="petal length (cm)",data=full_data)
plt.subplot(2,2,4)
sns.violinplot(x="Classification",y="petal width (cm)",data=full_data)
Скрытая информация :: Авторизуйтесь для просмотра »
The
thinner part denotes that there is
less density whereas the
fatter part conveys
higher density.
And similarly, we may use
boxplot to see how the
categorical feature Classification is distributed with all other input and also, to check for Outliers variables:
plt.figure(figsize=(12,10))
plt.subplot(2,2,1)
sns.boxplot(x="Classification",y="sepal length (cm)",data=full_data)
plt.subplot(2,2,2)
sns.boxplot(x="Classification",y="sepal width (cm)",data=full_data)
plt.subplot(2,2,3)
sns.boxplot(x="Classification",y="petal length (cm)",data=full_data)
plt.subplot(2,2,4)
sns.boxplot(x="Classification",y="petal width (cm)",data=full_data)

To check Cardinality:
def count_unique_values(theData, categorical_columns_list):
cats = theData[categorical_columns_list]
rValue = pd.DataFrame({'cardinality': cats.nunique()})
return rValue
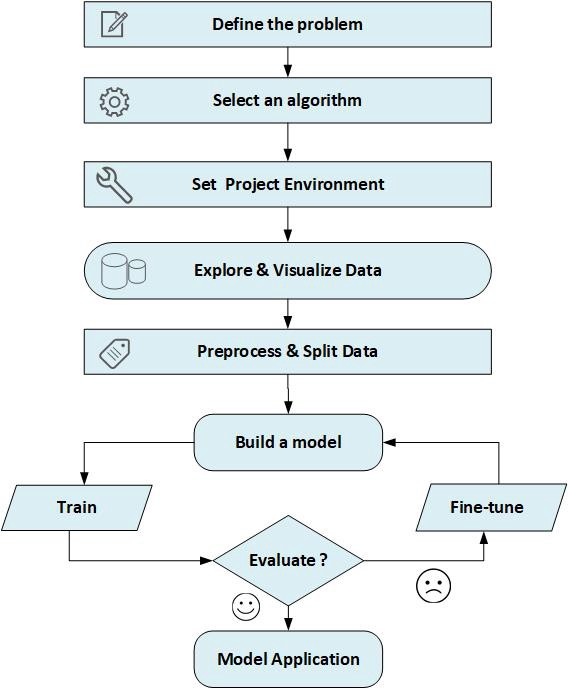
 Splitting Dataset Into Training and Testing Data
Splitting Dataset Into Training and Testing Data
We cannot use the same data we used to build the model to evaluate it. This is because our model can always simply remember the whole training set, and will therefore always predict the correct label for any point in the training set.
This
remembering does not indicate to us whether our model will generalize well, i.e., whether it will also perform well on new data.
So, Before using a machine learning model that can predict from unseen data, we should have some way to know whether it actually works or not.
Hence, we need to split the labeled data into two parts.
One part of the data is used to build our machine learning model, and is called the training data or
training set. The rest of the data will be used to measure how well the model works; this is called the test data, or
test set.
scikit-learn contains a function that
shuffles the dataset and
splits it for you: the
train_test_split function. This function extracts
75% of the rows in the data as the training set, together with the corresponding labels for this data.
The remaining
25% of the data, together with the remaining labels, is declared as the test set.
In scikit-learn, data is usually denoted with a capital X, while labels are denoted by a lowercase y.
This is inspired by the standard formulation f(x)=y in mathematics, where x is the input to a function and y is the output.
Following more conventions from mathematics, we use a capital X because the data is a two-dimensional array (a matrix) and a lowercase y because the target is a one-dimensional array (a vector). Let’s call train_test_split on our data and assign the outputs using this following code:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test =
train_test_split(mydata,Labels, random_state=0)print("X_train shape: {}".format(X_train.shape))
print("y_train shape: {}".format(y_train.shape))
print("X_test shape: {}".format(X_test.shape))
print("y_test shape: {}".format(y_test.shape))
Before making the split, the train_test_split function shuffles the dataset using a pseudorandom number generator.
If we just took the last 25% of the data as a test set, all the data points would have the label 2, as the data points are sorted by the label (see the output for iris[‘target’] shown earlier).
Using a test set containing only one of the three classes would not tell us much about how well our model generalizes, so we shuffle our data to make sure the test data contains data from all classes.
To make sure that we will get the same output if we run the same function several times, we provide the pseudo random number generator with a fixed seed using the random_state parameter.
This will make the outcome deterministic, so this line will always have the same outcome. The output of the train_test_split function is X_train, X_test, y_train, and y_test, which are all NumPy arrays. X_train contains 75% of the rows of the dataset, and X_test contains the remaining 25%.
Build the Model
Now we can start building the actual machine learning model. There are many classification algorithms in scikit-learn that we could use. Here, we will use a
k-nearest neighbors classifier, which is easy to understand. Building this model only consists of storing the training set.
To make a prediction for a new data point, the algorithm finds the point in the training set that is closest to the new point. Then it assigns the label of this training point to the new data point.
The k in k-nearest neighbors signifies that instead of using only the closest neighbor to the new data point, we can consider any fixed number k of neighbors in the training (for example, the closest three or five neighbors). Then, we can make a prediction using the majority class among these neighbors.
For simplification, we’ll use only a single neighbor.
All machine learning models in scikit-learn are implemented in their own classes, which are called Estimator classes. The
k-nearest neighbors classification algorithm is implemented in the
KNeighborsClassifier class in the neighbors module. Before we can use the model, we need to instantiate the class into an object. This is when we will set any parameters of the model.
The most important parameter of KNeighbor sClassifier is the number of neighbors, which we will set to 1:
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=1)
The knnobject encapsulates the algorithm that will be used to build the model from the training data, as well the algorithm to make predictions on new data points. It will also hold the information that the algorithm has extracted from the training data. In the case of
KNeighborsClassifier, it will just store the training set.
To build the model on the training set, we call the fit method of the knn object, which takes as arguments the NumPyarray X_train containing the training data and the NumPyarray y_train of the corresponding training labels:
knn.fit(X_train, y_train)
The
fit method returns the knn object itself (and modifies it in place), so we get a string representation of our classifier.
The representation shows us which parameters were used in creating the model.
Nearly all of them are the default values, but you can also find n_neighbors=1, which is the parameter that we passed.
Most models in scikit-learn have many parameters, but the majority of them are either speed optimizations or for very special use cases.
You don’t have to worry about the other parameters shown in this representation. Printing a scikit-learn model can yield very long strings, but don’t be intimidated by these. So, we will not show the output of fit because it doesn’t contain any new information.
We can now make predictions using this model on new data for which we might not know the correct labels.